Lineage of the Y-chromosome: where did dads come from?
For Father's Day, let's look at the boy genes
The Y chromosome is much smaller than the X chromosome. But an important gene on the Y chromosome includes: SRY, The SRY (Sex Determining Y Region) gene which creates the male phenotype.
Somatic chromosomes come in duplicates, and females have two X chromosomes. But the Y chromosome exists solely. When it mutates there is no backup, the gene will disappear. Over time the errors lead to elimination of the genes on the Y chromosome. It now has only ~ 80 genes.
However, the Y chromosome makes mirror-image copies of its most crucial genes – palindromes. These contain genetic information reading forward in the first half, and then the same information is repeated in reverse, so the most important genes on the Y chromosome do occur in tandem, but on the same chromosome.
Determining Y Chromosome’s Past
Lots has been written about our genetic “Eve” as derived from the data analysis of mitochondrial DNA. But less is known about the source of “Adam” info. This comes from sequencing the Y chromosome across global populations.
The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve" (mt-MRCA, the matrilineal transmission of mtDNA), the most recent woman from whom all living humans are descended matrilineally.
Paternal lineage DNA tests can trace a family’s origins through the Y chromosome. This test works for males, as the DNA handed down on the Y chromosome is nearly identical to the man’s ancestor’s DNA. The DNA on the Y chromosome changes little, so a man can trace his family ancestry through genetic testing of his own Y chromosome.
The chromosome is tested for markers in loci on the Y chromosome. These markers are numbered 1-33 or 1-46. The number assigned to each is equal to the number of “short tandem repeats” (STRs) at the specific genetic location. When a man compares the number of STRs for each marker, he can find distant relatives who share the same DNA. In addition, the Y chromosome belongs to a specific haplogroup, that can point to past family origins.
From L Lefler:
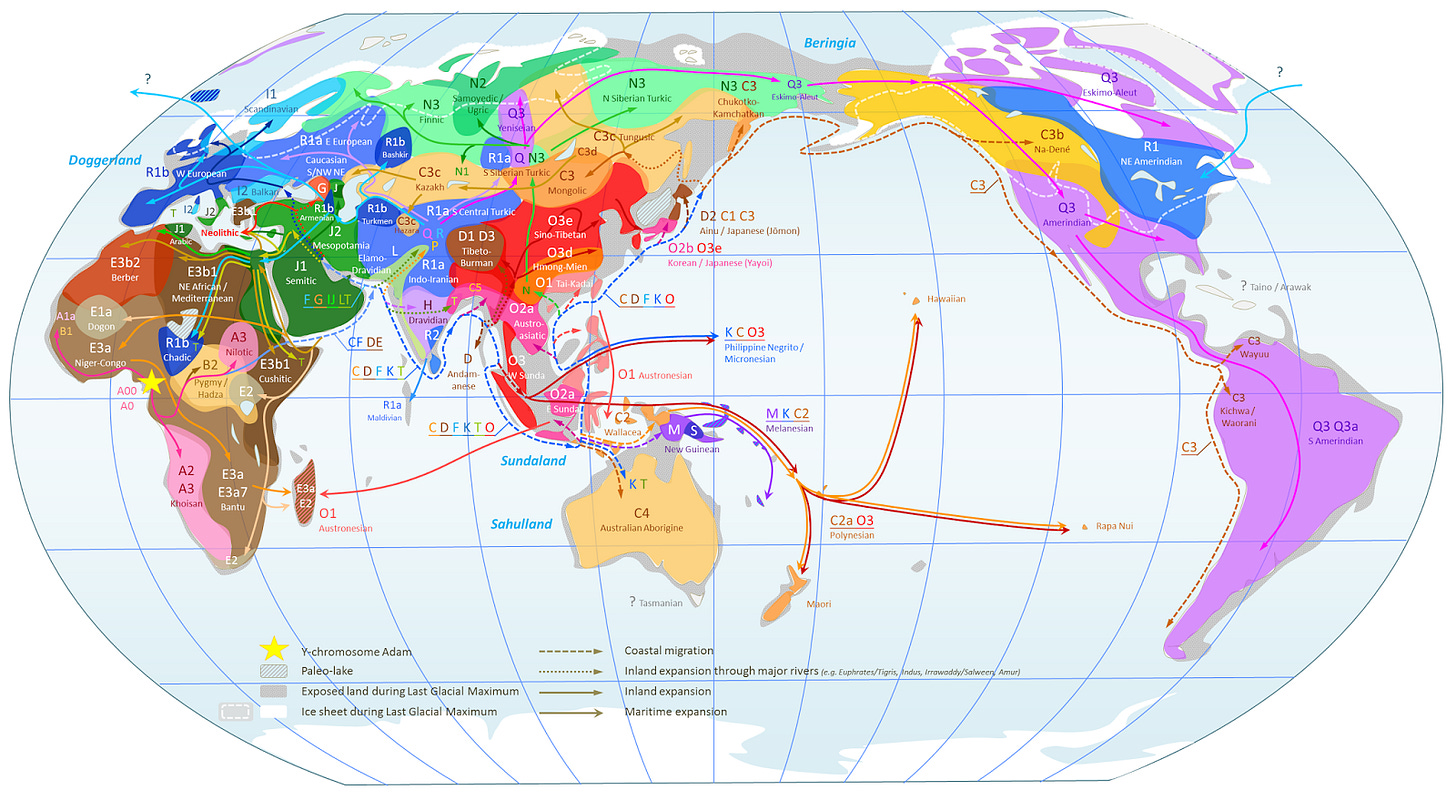
Haplogroup A: One of the oldest haplogroups. Found in Southern Africa and the Southern Nile region.
Haplogroup B: This haplogroup is only found in Africa and is one of the oldest known haplogroups.
Haplogroup C: This group colonized Australia, New Guinea, and is also found in India and among some Native American tribes. The C group started in southern Asia and spread in many directions.
Haplogroup D: The first Japanese humans originated from this haplogroup, which began in Asia. The aboriginal Japanese still carry this haplogroup, along with inhabitants of the Tibetan plateau.
Haplogroup E: Haplogroup E is only found in Africa. Interestingly, this haplogroup is closely related to Haplogroup D, which is not found in Africa. This haplogroup arose in Northeast Africa or the Middle East.
Haplogroup F: This haplogroup is the "father" of most of the remaining haplogroups - haplogroup F is the originator of haplogroups G through R. Over 90% of the world's population comes from this haplogroup or one of its progeny. This haplogroup originated during the migration out of Africa: it is almost never found within sub-Saharan Africa.
Haplogroup G: Originating from Pakistan or India, this haplogroup is found in Europe, central Asia, and the Middle East.
Haplogroup H: This haplogroup is only found in Sri Lanka, India, and Pakistan.
Haplogroup I: Nearly one-fifth of the European population carries this haplogroup. This haplogroup is not found outside of Europe and probably arose when ice sheets covered most of Europe. Certain subtypes are specific to Scandinavians and those from Iceland. Viking invaders and Anglo-Saxons can be typed from the British population using specific markers from this haplogroup.
Haplogroup J: North Africa and the Middle East are the locations where haplotype J is found - it probably arose in this general geographic area.
Haplogroup K: Haplogroup K is found in Iran and the southern region of Central Asia. It is estimated that this haplogroup arose 40,000 years ago.
Haplogroup L: Sri Lankans, people from India, and others from the Middle East may carry haplogroup L.
Haplogroup M: A whopping 33%-66% of the Papua, New Guinea population carries haplogroup M.
Haplogroup N: While this haplogroup probably arose in China or Mongolia, it is currently found in Northeast Europe and Siberia. This is the most common haplogroup among Finns and native Siberians.
Haplogroup O: The vast majority (80%) of East Asians are haplogroup O. It evolved about 35,000 years ago and is found exclusively in East Asia.
Haplogroup P: This is a fairly rare haplogroup found in India, Pakistan, and central Asia.
Haplogroup Q: Native Americans and Northern Asians carry haplotype Q: this is the population that migrated out of Asia and into North America. Native Americans carry the specific haplotype Q3, which is restricted to the population of humans who migrated across the Bering Strait.
Haplogroup R: One strain of this haplogroup is found among people who live near the Caspian Sea in Eastern Europe: the culture of the people in this area domesticated the first horses. Another haplotype from the R group is found in Europe and is found in high concentrations in Western Ireland.
Haplogroup S: The highlands of Papua, New Guinea carry this haplogroup, along with some people living in nearby Indonesia and Melanesia.
Haplogroup T: This haplogroup is found in Southern Europe, Southwest Asia, and throughout Africa.
World Map of Y-Chromosome Haplogroups: Dominant Haplogroups in Pre-Colonial Populations with Possible Migrations Routes
Lifestyle Affects Mutation Rate
A team of researchers from France and Germany analyzed more than 500 sequences of the male Y- chromosome in southern African ethnic groups who are farmers and in population groups engaged in traditional hunter-gatherer activities. The agriculturalists had a higher rate of change when compared to hunter-gatherers. This is attributed to significantly older average age of paternity among the farmers.
Therefore the Y chromosome, changes at different speeds in different population groups. Farmers marry more than one wife, siring children at advanced ages with younger women. Men from the farming societies had progeny longer, had an older average age for fatherhood. Their mutation rate was higher than for men from foraging societies. A 15-year increase in age of paternity resulted in a 50% increase in chromosome mutations.
This Fathers Day, while grilling your roast beast, along side the recently harvested zucchini, savor the long ago legacies scripted into your genes.
REFERENCES
The Y Chromosome: The Why Factor. How the Y chromosome transforms a female embryo to a male one and the differences between men and women. https://www.bbc.co.uk/programmes/p02rf5qh https://www.bbc.co.uk/sounds/play/p02rrc68
L Lefler. The Y Chromosome: Ancestry, Genetics, and the Making of a Man. JUL 27, 2022. https://owlcation.com/stem/The-Y-Chromosome-Ancestry-Genetics-and-the-Making-of-a-Man
Y-chromosomal Adam. https://en.wikipedia.org/wiki/Y-chromosomal_Adam
C Barbieri, et al. Refining the Y chromosome phylogeny with southern African sequences Human Genetics (April 2016) doi:dx.doi.org/10.1101/034983
Genetics reveals the impact of lifestyle on evolution (2016, April 4) https://medicalxpress.com/news/2016-04-genetics-reveals-impact-lifestyle-evolution.html








Fascinating!